
सीएसआईआर-केन्द्रीय औषधीय एवं सगंध पौधा संस्थान
CSIR-Central Institute of Medicinal and Aromatic Plants (CSIR-CIMAP)




Harnessing Artificial Intelligence for Natural Products Discovery
Welcome to the AI-Lab, led by Dr. Aman Chandra Kaushik, where we integrate bioinformatics, cheminformatics, and machine learning to accelerate natural product research and plant-microbe interaction studies.
 Dr. Aman Kaushik – Bioinformatician at CIMAP
Dr. Aman Kaushik – Bioinformatician at CIMAP
Dr. Aman Kaushik is a Scientist and Bioinformatician at Central Institute of Medicinal and Aromatic Plants (CIMAP), CSIR, India.
He specializes in integrating computational biology, cheminformatics, and artificial intelligence to drive innovations in plant genomics, natural products research, and molecular systems biology.
Currently, he is leading cutting-edge initiatives under the The Ayushman Bharat Digital Mission vision, targeting AI-powered agri-nutrition-biotech transformations for sustainable healthcare and agriculture.
 AI-driven analysis of plant molecules
AI-driven analysis of plant molecules
 AI lab interface for plant compounds
AI lab interface for plant compounds
 Intelligent discovery of plant metabolites
Intelligent discovery of plant metabolites
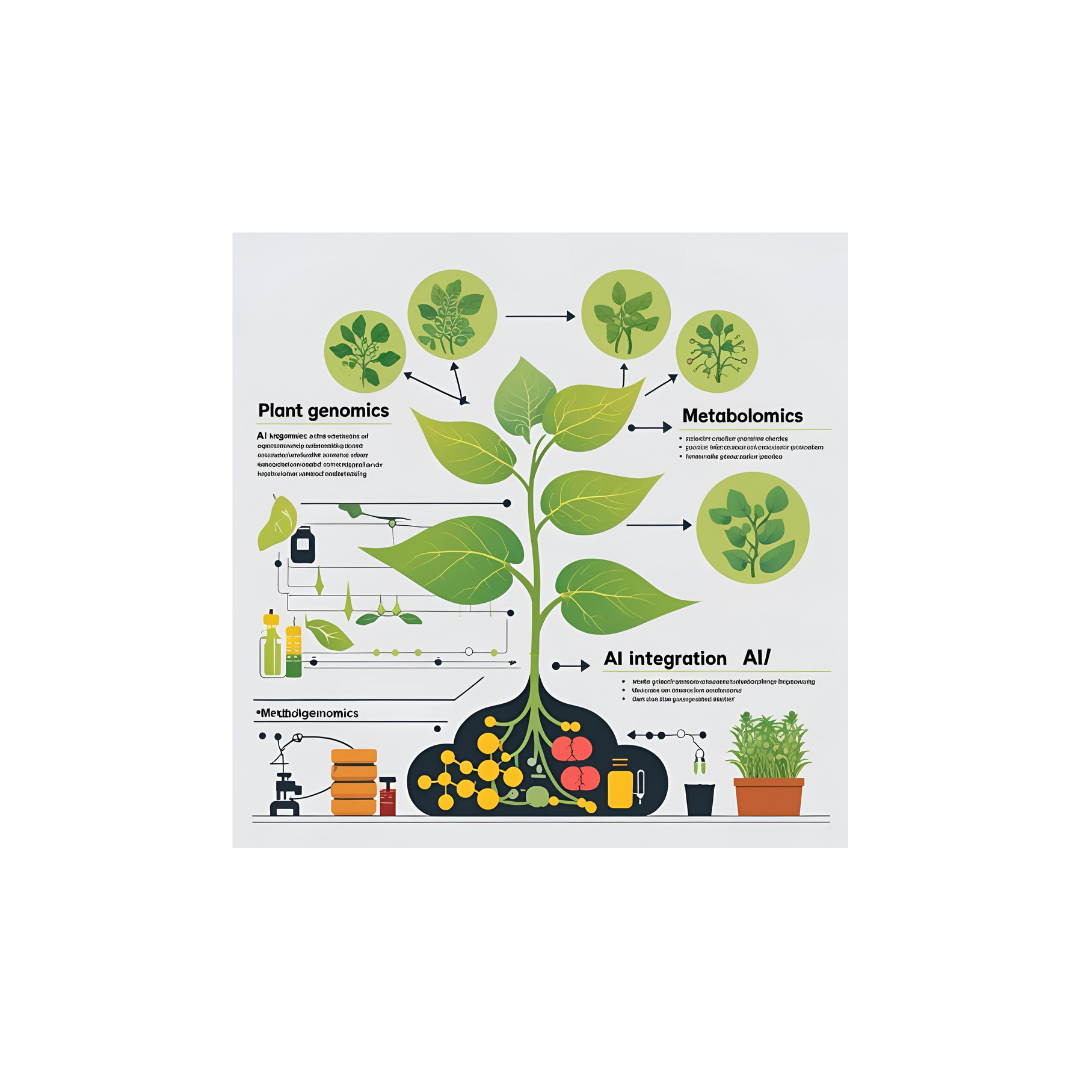 Plant genomics × AI × Metabolomics
Plant genomics × AI × Metabolomics
Welcome to the portfolio of computational tools and pipelines developed to facilitate advanced bioinformatics analyses, particularly focusing on plant and microbial genomics, transcriptomics, and metabolomics.
These solutions are designed with dashboard-style interfaces, integrating AI models and domain-specific visualizations to help researchers intuitively explore and interpret complex biological data.
These tools integrate advanced AI models and computational algorithms, illustrated by neural network architectures overlaid with plant metabolic pathways to highlight the synergy between machine learning and biological insight.
Our platforms aim to empower researchers with interactive, visually-rich environments to accelerate discoveries in plant science and bioinformatics.
